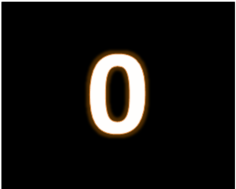In two recently released analysis we demonstrated a never before achieved reduction of false positives by use of our product ReSurfX::vysen on a data that has been extensively studied to develop numerous analytic technologies. That data is from large-scale gene expression technology analysis – Read on.. don’t let that field of application sway your interest. This never before possible level of accuracy was made possible by a breakthrough data-source agnostic 'Adaptive Hypersurface Technology' (AHT) that ReSurfX is leveraging in data-analytics and continuing to develop. Here, we discuss the state-of-the-art in data-analytics, current thoughts and problems (from popular media and authoritative source). What does it mean for your innovation and ROI?
Read more...
How can ZERO get a great ROI for your enterprise and improve your innovation potential? Big Data brings more opportunities and errors into your workflow. Robust accuracy and automatable knowledge extraction are keys to successfully leveraging this digital transformation.
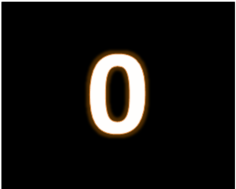 Recently we showed how a ZERO from the ReSurfX::vysen product should yield a great ROI and improve innovation for your enterprise using an extensive analysis of sequencing (RNASeq). Here we repeat that excercise with Microarray data and show that the UNPRECEDENTED VALUE OBTAINED FOR RNAseq DATA IS REPLICATED to this platform as well. This is of incredible importance, given the data-source agnostic property of Adaptive Hypersurface Technology (AHT).
Recently we showed how a ZERO from the ReSurfX::vysen product should yield a great ROI and improve innovation for your enterprise using an extensive analysis of sequencing (RNASeq). Here we repeat that excercise with Microarray data and show that the UNPRECEDENTED VALUE OBTAINED FOR RNAseq DATA IS REPLICATED to this platform as well. This is of incredible importance, given the data-source agnostic property of Adaptive Hypersurface Technology (AHT).
Recently we released a new version of our analytics product ReSurfX::vysen 2.0 – showcasing the power of our Adaptive Hypersurface Technology (AHT)™ using RNAseq and Microarray based gene expression analysis.
As we had done previously with sequencing quality control consortium (SEQC) benchmark data (RNAseq), here we carried out a similar analysis using microarray quality control consortium (MAQC) data from two sites that had each carried out analysis on the same two tissue samples with 5 replicates each. We AGAIN found that ReSurfX:: vysen:
- Identified ‘ZERO’ false positive differentially expressed gene from about 10 million calls IN 180 (EVERY within-sample 3-replicate) COMPARISON EACH INVOLVING 54,675 GENES, proving remarkable false positive control.
- Is incredibly MORE SENSITIVE AND REPRODUCIBLE THAN ANY OTHER ANALYTICS APPROACH comparing ALL POSSIBLE between-sample THREE-REPLICATE COMBINATIONS from each of the two sites. For this purpose vysen was used in over 11 million calls for differentially expressed genes across 200 comparisons.
The strength of these results from vysen are AGAIN unprecedented with enormous impact for analytics and outcomes highlighting the versatility of the data-source agnostic AHT technology despite very different raw data properties of Microarray and RNAseq data. Here we share the details of this unprecedented accuracy.
Read more...
How can a ZERO from an analytics product get a great ROI and improve innovation for your enterprise?
R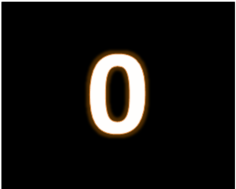 ecently we released a new version of our analytics product ReSurfX::vysen 2.0 – initially showcasing the power of our Adaptive Hypersurface Technology (AHT)™ using RNAseq and Microarray based gene expression analysis.
ecently we released a new version of our analytics product ReSurfX::vysen 2.0 – initially showcasing the power of our Adaptive Hypersurface Technology (AHT)™ using RNAseq and Microarray based gene expression analysis.
Here we prove the power of vysen using unprecedented results with enormous impact for analytics and outcomes. This forms one basis for vysen being the most accurate product in the market.
We analyzed sequencing quality control consortium (SEQC) benchmark data from two sites (Mayo Clinic and Beijing Genomics Institute) that had each carried out sequencing (RNAseq) analysis on two different tissue samples with 5 replicates each. We found that ReSurfX::vysen :
- Identified ‘ZERO’ false positive differentially expressed gene from over 10 million calls IN 180 (EVERY within-sample 3-replicate) COMPARISONS EACH INVOLVING 58,051 GENES, proving remarkable false positive control.
- Is incredibly MORE SENSITIVE AND REPRODUCIBLE THAN ANY OTHER APPROACH through direct comparison, as well as comparing ALL POSSIBLE between-sample THREE-REPLICATE COMBINATIONS. This involved 200 vysen comparisons of 58,051 genes in over 11 million DEG calls.
Read more...