Unprecedented value of ReSurfX::vysen produced “ZERO” in Sequencing & Microarray data: a case of Goose & Gander?
How can ZERO get a great ROI for your enterprise and improve your innovation potential? Big Data brings more opportunities and errors into your workflow. Robust accuracy and automatable knowledge extraction are keys to successfully leveraging this digital transformation.
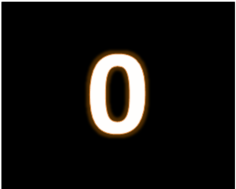 Recently we showed how a ZERO from the ReSurfX::vysen product should yield a great ROI and improve innovation for your enterprise using an extensive analysis of sequencing (RNASeq). Here we repeat that excercise with Microarray data and show that the UNPRECEDENTED VALUE OBTAINED FOR RNAseq DATA IS REPLICATED to this platform as well. This is of incredible importance, given the data-source agnostic property of Adaptive Hypersurface Technology (AHT).
Recently we showed how a ZERO from the ReSurfX::vysen product should yield a great ROI and improve innovation for your enterprise using an extensive analysis of sequencing (RNASeq). Here we repeat that excercise with Microarray data and show that the UNPRECEDENTED VALUE OBTAINED FOR RNAseq DATA IS REPLICATED to this platform as well. This is of incredible importance, given the data-source agnostic property of Adaptive Hypersurface Technology (AHT).
Recently we released a new version of our analytics product ReSurfX::vysen 2.0 – showcasing the power of our Adaptive Hypersurface Technology (AHT)™ using RNAseq and Microarray based gene expression analysis.
As we had done previously with sequencing quality control consortium (SEQC) benchmark data (RNAseq), here we carried out a similar analysis using microarray quality control consortium (MAQC) data from two sites that had each carried out analysis on the same two tissue samples with 5 replicates each. We AGAIN found that ReSurfX:: vysen:
- Identified ‘ZERO’ false positive differentially expressed gene from about 10 million calls IN 180 (EVERY within-sample 3-replicate) COMPARISON EACH INVOLVING 54,675 GENES, proving remarkable false positive control.
- Is incredibly MORE SENSITIVE AND REPRODUCIBLE THAN ANY OTHER ANALYTICS APPROACH comparing ALL POSSIBLE between-sample THREE-REPLICATE COMBINATIONS from each of the two sites. For this purpose vysen was used in over 11 million calls for differentially expressed genes across 200 comparisons.
The strength of these results from vysen are AGAIN unprecedented with enormous impact for analytics and outcomes highlighting the versatility of the data-source agnostic AHT technology despite very different raw data properties of Microarray and RNAseq data. Here we share the details of this unprecedented accuracy.
[Learn more about ReSurfX:: vysen here!]
We took part of the dataset from MAQC (Microarray Quality Control Consortium) that made the most sense for real-life applications i.e., comparing gene expression data in two samples [the Universal Human Reference RNA (UHRR, from 10 pooled cancer cell lines, Agilent Technologies, Inc.) and the Human Brain Reference RNA (HBRR, from multiple brain regions of 23 donors, Life Technologies, Inc.] – or as the samples were labelled by the consortium: "A vs B". The analytics and outcomes presented below can be reproduced from our trial version of the product (soon to be incorporated into the commercial version).
We took data from two sites in the study (Site1 and Site2) generated by the Affymetrix™ GeneChip® platform. The data was generated at multiple sites to compare reproducibility and study variability. Each of the sites carried out GeneChip® analysis of FIVE technical replicates of each of the two samples.
First, some salient and unique features of ReSurfX:: vysen output:
- ReSurfX::vysen gives a 0 vs 1 for No Change vs Change, respectively between two conditions being compared (removing the need for a subjective p-value, fdr-thresholding).
- ReSurfX::vysen identifies significantly more overlapping DEGs between the sites than other analytics workflows used, as seen by comparing samples AvsB (Fig. 1). Thus fact can be seen in numerous publications to date that tried to get the level of false positives to less than 10% by various approaches.
- ReSurfX::vysen has an automatable outlier detection system that will be available in the product in the future.
- Interestingly, the ReSReSurfX::vysen performs extremely well even with a limited number of replicates (which is a rarity in available analytic approaches).
Summary of ReSurfX::vysen analytics and the power of our product and technology:
- Incredibly, when we took all possible within-sample three-replicate comparisons from both sites for both samples, ReSurfX::vysen produced ‘ZERO’ differentially expressed genes – IN EVERY 180 COMPARISONS EACH INVOLVING 54,675 GENES. No analytic result has been reported to-date with this level of false positive control.
- Given the enormous implication of finding ZERO within-sample false positive DEGs, we went on to study all combinations of between-sample three-replicates from Site1-SampleA vs Site1-SampleB. We could reproducibly identify the same genes (error rate less than 3% as ahown in Fig.2 and Fig.3) with extreme sensitivity in three replicate comparisons between the two tissues at both the sites. In principle (because we got ZERO DEG in 10 million calls across 180 within-sample comparisons) in the analysis of within-sample data, if these results are reproducibly found between all these comparisons - then it is reasonable to conclude that the DEGs identified in between-sample comparisons are more likely to represent true positives.
a. We compared ALL POSSIBLE between-sample THREE REPLICATE COMBINATIONS from each of the sites.
b. We found that the number of differentially expressed genes (DEGs) identified AND the overlap among them was INCREDIBLY consistent. Further, the number of DEGs identified was significantly larger than had been reported by any other study, indicating that ReSurfX::vysen identifies significantly more DEGs with added sensitivity and reproducibility despite the level of false positive reduction shown above . THIS SHOWS that vysen is INDEED, MORE SENSITIVE THAN ANY OTHER ANALYTIC APPROACHES AVAILABLE, EVEN AFTER EXTENSIVE DEVELOPMENT OF A VARIETY OF ANALYTIC APPROACHES BY THE FIELD.
c. Comparing the outcome of this analysis as in point #2 above indicates THE ROBUST POWER OF ReSurfX::vysen TO PERFORM WELL EVEN IN THE PRESENCE OF SOME OUTLIERS. This can be seen from Fig. 1 that identifies a lot more DEGs than any reported study that has controlled for false positives. - These results are consistent with our claim and value proposition – we can get more accurate results than other analytic methods available in the market and sector with FEWER replicates/subjects!
- Even with additional filters like widely practiced 2-fold change threshold of real-life datasets and outcomes, this level of accuracy and reproducibility has never been reported. Thus, Adaptive Hypersurface technology (AHT) – should be an IMMENSELY POWERFUL addition to any analytics tool box to reap value in an increasingly data-centric world.
NOTE: Data from the MAQC database was not used as a training set for analytics or technology development of the ReSurfX::vysen product at any point (so there is no inherent or specific overfitting in the technology and product to the data presented here).
Outline of analysis details: [WE INVITE YOU TO COMPARE YOUR RESULT AND SHARE IT WITH THE WORLD AS COMMENT HERE]
MAQC data (GSE5350; Nat Biotechnol 2006 Sep;24(9):1151-61) representing the 5 replicates of the Universal Human Reference RNA (UHRR, from 10 pooled cancer cell lines, Agilent Technologies, Inc.) and the Human Brain Reference RNA (HBRR, from multiple brain regions of 23 donors, Life Technologies, Inc.) were used.
The data were downloaded as CEL files from the NCBI/GEO database. The resulting sample reads in CEL format files were processed using vysen using signal using in-built normalization developed by ReSurfX. The format of the data files were converted to suit the AHT workflow before comparative analytics using AHT.
Quantitative support (click on images to see them at higher resolution):
Fig. 1: Overlap of Site1 vs Site2 sample AvsB comparisons. 5 replicates each Sample A vs Sample B from Site1 (right circle) vs Site2 (left circle).
Note, in all outputs shown below – no external thresholding (like fold change, p-value, fdr were applied – instead, ReSurfX::vysen calls of 1 or 0 for DEG was used. vysen also gives a confidence value (that makes sense internally in a comparison and in conjunction with a DGE call).
Fig.2: Number of DGEs found in each of the 180 possible 3-replicate within-sample comparisons in each of the two samples from each of the two sites. [Total 180 comparisons of 54,675 genes].
Remarkably, no DGEs were detected by ReSurfX::vysen in all 180 comparisons.
Fig. 3: Remarkable consistency among all possible 3-replicate comparison between Sample A and Sample B from Site1. [100 comparisons]
Shown in the graph are the number of comparisons that called the same gene as DEGs (Count) out of maximum possible 100. Note: 54,675 genes in all cases. 100 implies all possible comparisons called a gene DEG, 0 implies all possible comparisons called the gene non-DEG).
The table shows the calculated of total true and false calls as explained in the previous post: The table shows average DEG calls over 100 comparisons of 54,675 genes, the maximum and minimum DEGs found in the 100 comparisons that also shows the spread is not very much (i.e., difference between maximum and minimum number of DEG are very low. The table also shows total TRUE POSITIVES, ESTIMATED FALSE NEGATIVES AND ESTIMATED ERROR INVOLVING OVER 5 MILLION CALLS. One additional observation is that there are a few DGEs where the estimated ratio is very close to 1 (likely due to the model for estimating unified value). To give an estimate, the maximum count of non-uniform DEGs was 582 (of 54,675 genes) at 1 (of 100 comparisons). 13,470 and 35,298 genes were called identically as DEGs or non-DEGs across all 100 comparisons.
Fig 4: Remarkable consistency among all possible 3-replicate comparison between Sample A and Sample B of Site2. [100 comparisons]
Shown in the graph are the number of comparisons that called the same gene as DEGs (Count) out of maximum possible 100. Note: 54,675 genes in all cases. 100 implies all possible comparisons called a gene DEG, 0 implies all possible comparisons called the gene non-DEG).
The table shows the calculated of total true and false calls as explained in the previous post: The table shows average DEG calls over 100 comparisons of 54,675 genes, the maximum and minimum DEGs found in the 100 comparisons that also shows the spread is not very much (i.e., difference between maximum and minimum number of DEG are very low. The table also shows total TRUE POSITIVES, ESTIMATED FALSE NEGATIVES AND ESTIMATED ERROR INVOLVING OVER 5 MILLION CALLS. One additional observation is that there are a few DGEs where the estimated ratio is very close to 1 (likely due to the model for estimating unified value). To give an estimate, the maximum count of non-uniform DEGs was 791 (of 54,675 genes) at 1 (of 100 comparisons). 14,144 and 32,540 genes were called identically as DEGs or non-DEGs across all 100 comparisons.
We also checked if the most used ratio cut-off (of 2.0) improved the results these from Site1 using ReSurfX::vysen. This resulted in far less DEGs (9,061) uniformly across 100 comparisons and 37,035 genes uniformly called non-DGEs. This is the most accurate result published that we are aware of with this level of false positive control (i.e., ZERO).
BACK TO YOUR ROI: [Learn more about ReSurfX:: vysen here!]
- ReSurfX::vysen find differentials accurately and reproducibly, improving: 1) the value of your data analytics efforts, 2) manpower and other resource utilization, 3) time to market with a correct product, and 4) outcomes of your business or research interest.
- ReSurfX::vysen has been previously used at a customer site. It did not find differentials in comparative analytics when they were not present with reliable analytic accuracy/reproducibility, as confirmed by a variety of other analytics methods.
REITERATING OTHER VALUE PROPOSITIONS OF ReSurfX::vysen:
- Extremely high accuracy, which by itself leads to novel insight due to high accuracy data analysis in your workflow.
- Highly reproducible on a variety of datasets – enabling automation of knowledge extraction from large volumes of data.
- Data-source agnostic AHT is applicable to a large variety of problems to analyze data from every source and in combinations for integrative analysis using the same technological principle and computational modules we have developed.
- Solutions end-to-end in your enterprise – data generation – discovery – clinical trials – outcomes/value-based product sale and patient outcomes – quantitative support for evidence based medicine – inherent personalization in outcomes prediction – Advance Outcomes Alert System™ (AOAS™).





BREAKTHROUGH levels of false positive reduction: what does it mean for your innovation and ROI | ReSurfX, Oct 03, 2017
[…] (SEQC & MAQC) generated using RNAseq and another microarray technology vysen identified ‘ZERO’ false positive differentially expressed gene from about 20 million calls in 360 comparisons each involving over […]